Junichi Tokuda, PhD, and Nobuhiko Hata, PhD, both of the National Center for Image-Guided Therapy…
Researchers Hope to Unlock Brain’s Mysteries With New MAGNUS MRI Scanner
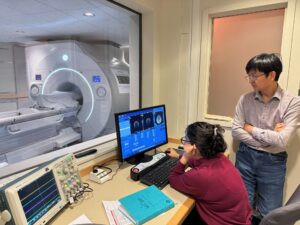 One of the world’s most advanced neuroimaging devices — able to reveal the fine structure of the brain with an unprecedented level of detail — has been installed at Brigham and Women’s Hospital.
One of the world’s most advanced neuroimaging devices — able to reveal the fine structure of the brain with an unprecedented level of detail — has been installed at Brigham and Women’s Hospital.
The Brigham is the fourth site in the world and the first in New England to acquire a MAGNUS scanner, an ultra-high-resolution MRI system that promises to reveal brain changes related to traumatic brain injury, Alzheimer’s disease, multiple sclerosis, brain tumors and many other neurological and psychiatric disorders. The Brigham system was delivered in late December and became operational in April. It was supported by a grant from the Massachusetts Life Sciences Center, with additional funding from Harvard Medical School and the hospital itself.
Source: Brigham Bulletin